Chapter 8 Human Pangenomes
8.1 Draft Human Pangenome Reference
8.1.1 Samples
47 phased, diploid genomes (aim for 350)
- 29 lymphoblastoid cell lines
- “limiting selection to those lines classified as karyotypically normal and with low passage (to avoid artefacts from cell culture)”
- 18 sequenced by others (some supplemented)
- Aimed for “genetic and biogeographic diversity”

Population_Code Description Super_Population_Code
ASW American’s of African Ancestry in SW USA AFR
ACB African Carribean in Barbados AFR
PUR Puerto Rican from Puerto Rica AMR
CLM Colombian from Medellian, Colombia AMR
PEL Peruvian from Lima, Peru AMR
MSL Mende in Sierra Leone AFR
GWD Gambian in Western Division AFR
YRI Yoruba in Ibadan, Nigera AFR
ESN Esan in Nigera AFR
MKK Maasai in Kinyawa, Kenya AFR
PJL Punjabi in Lahore, Pakistan SAS
CHS Southern Han Chinese EAS
KHV Kinh in Ho Chi Minh City, Vietnam EAS
Super-Populations
AFR, African
AMR, Ad Mixed American
EAS, East Asian
EUR, European
SAS, South Asian
8.1.2 Strategy
Sequencing
PacBio HiFi
Oxford Nanopore
Bionano optical maps
High-coverage Hi-C
Illumina short-read sequencing
High-coverage Illumina sequencing data for both parents
Assembly
Trio-HiFiasm
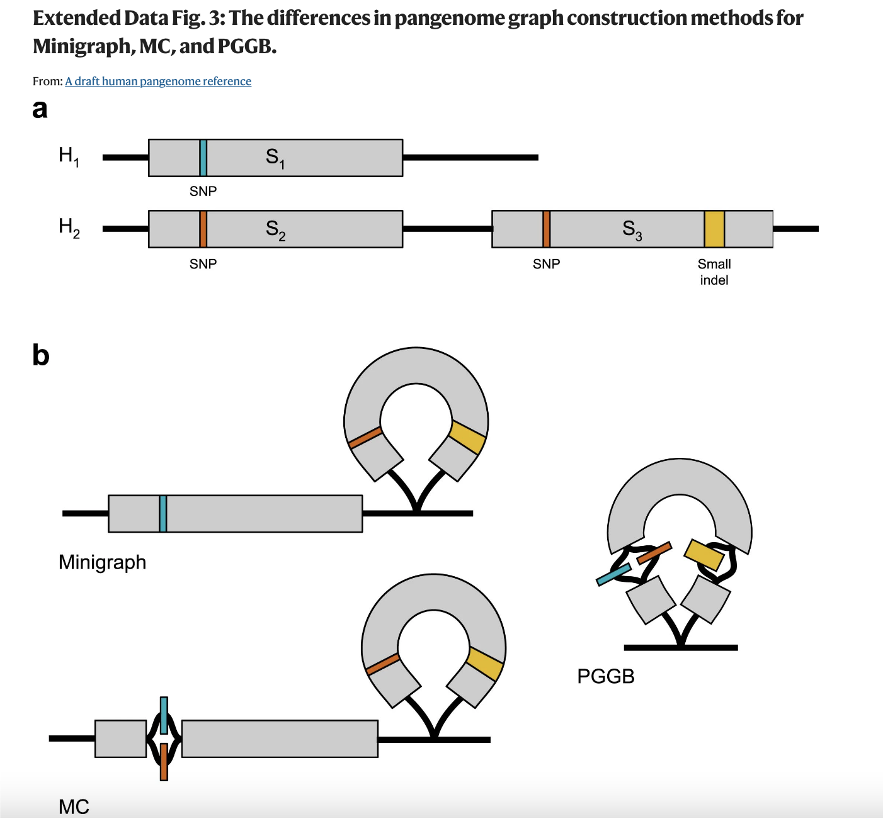
Graphs
Minigraph
* Fast pangenome graph builder based on the minimap2 aligner
* Only structural variation >=50nt
Minigraph-Cactus (MC)
* Refines minigraph output to include SNPs and other small variants
* Rewrote minigraph to write chains of minimizers
* Rewrote cactus to be able to read in minigraph output
PanGenome Graph Builder (PGGB)
* All pairwise genome assembly alignments -> graph
* Uses graph normalization to make sure that chromosome paths are linear
* Allows for cyclic graph structures that capture structural variation.
8.1.3 Results
More Genetic Variation Captured
The pangenome captures more polymorphic sequences
- 119 Mb of euchromatic polymorphic sequences
- 90 MB = structural variation
- 1,115 gene duplications
We can align more (short) reads to the pangenome
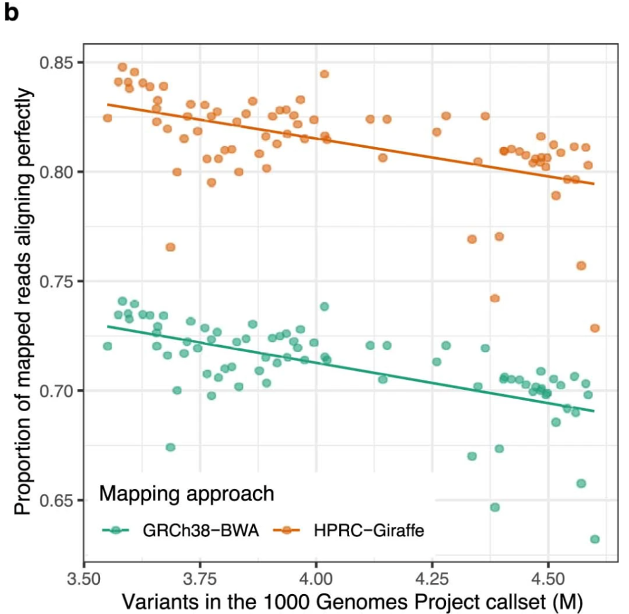
We can call more variants more accurately
Aligning short reads to the pangenome
- lowered error in small variants by 34%
- increased structural variants calls per haplotype by 104% (“vast majority”)
We can call variants across a broad set of populations

Variation in complex, medically-relevant Regions
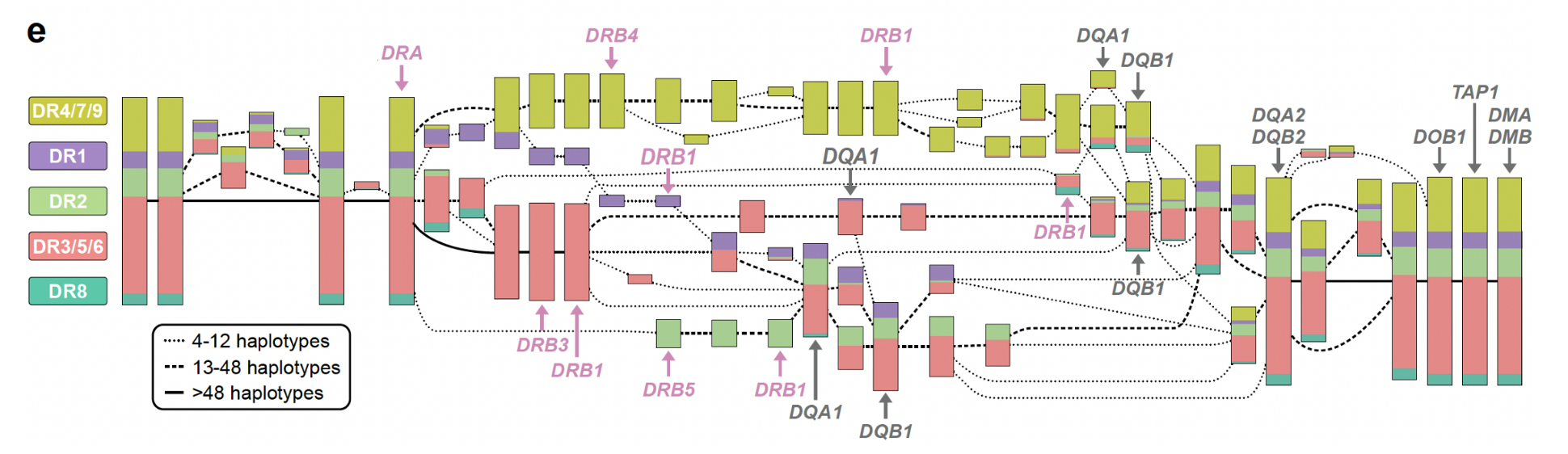
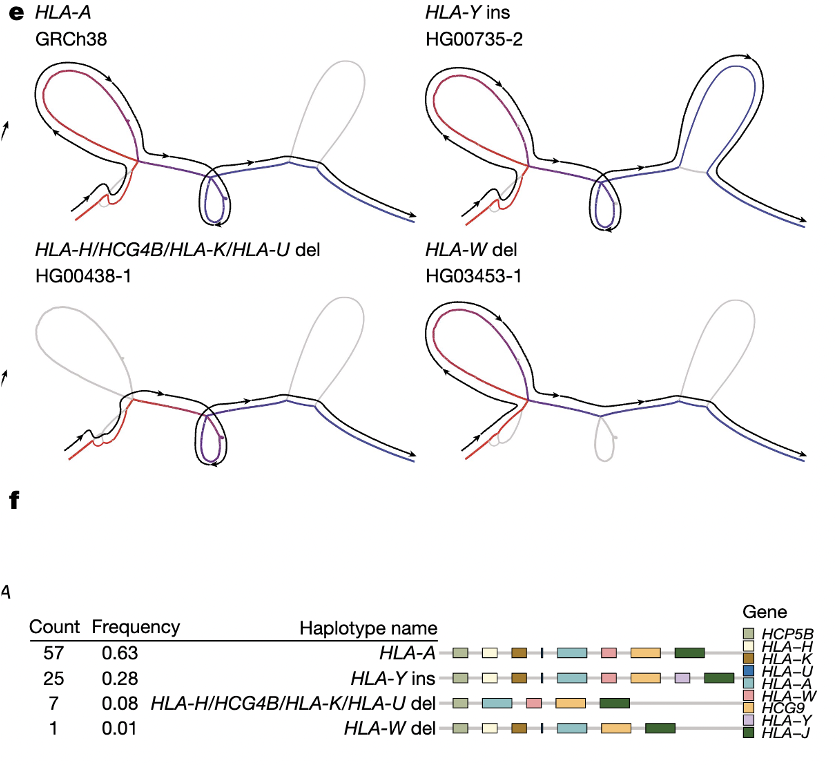
HLA region (helps the immune system distinguish between self and invader)

Rh region (involved in Rh blood type)

8.2 Complex Variation
8.2.1 Samples
65 phased, diploid human genomes
Closed 92% gaps from previous assemblies
Telomere-to-telomere (T2T) for 39% of chromosomes
8.2.3 Data Availability
https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/HGSVC3/release/