9 Nanopore WGS Community Profiling (KrakenUniq)
KrakenUniq
“confident and fast metagenomics classification using unique k-mer counts”
Default kmer = 31
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1568-0
https://github.com/fbreitwieser/krakenuniq
https://gitlab.umiacs.umd.edu/derek/krakenuniq/-/blob/master/MANUAL.md
9.1 Kraken/Centrifuge Family
Kraken 1 is obsolete.
Centrifuge was written to improve Kraken’s memory issues. It is completely new code with a different classification index. Its databases are also much smaller. Centrifuge can assign a sequence to multiple taxa. We’ll use centrifuge later in the workshop.
KrakenUniq is based on Kraken 1. It adds an efficient algorithm to assess coverage of unique kmers, running at the same speed and with only slightly more memory than Kraken 1. KrakenUniq distinguishes low abundance organisms from false positives.
Kraken 2 uses “probabilistic data structures” to reduce memory and increase speed at the expense of lower accuracy that leads to false positives during the classifications (“a few dozen out of millions”).
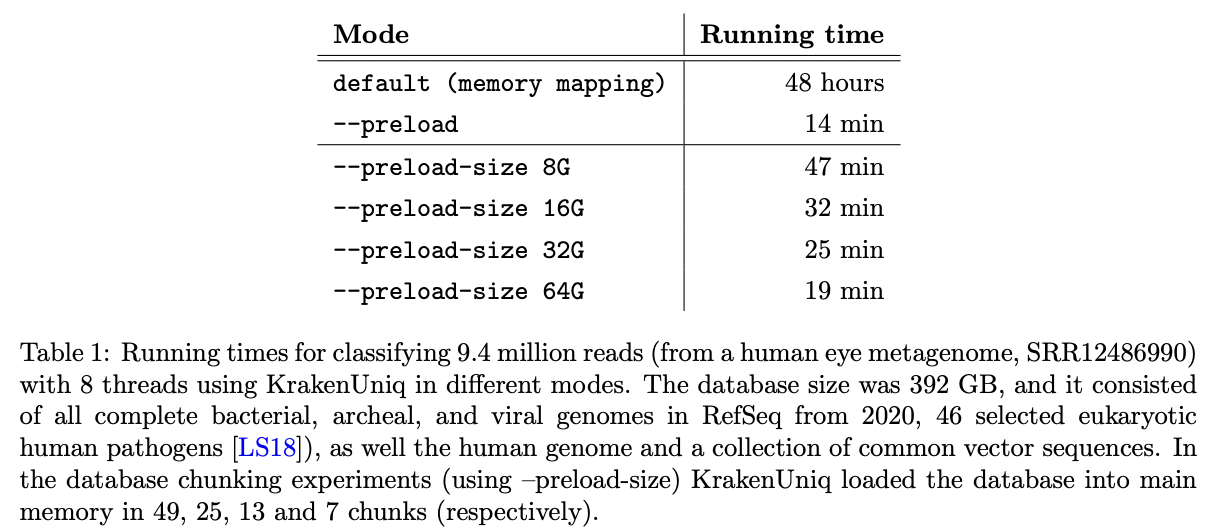
KrakenUniq was updated in May 2022 (v0.7+) to help to deal with the large databases on computers without enough RAM to load them into memory. Databases can be read in chunks that fit in RAM.
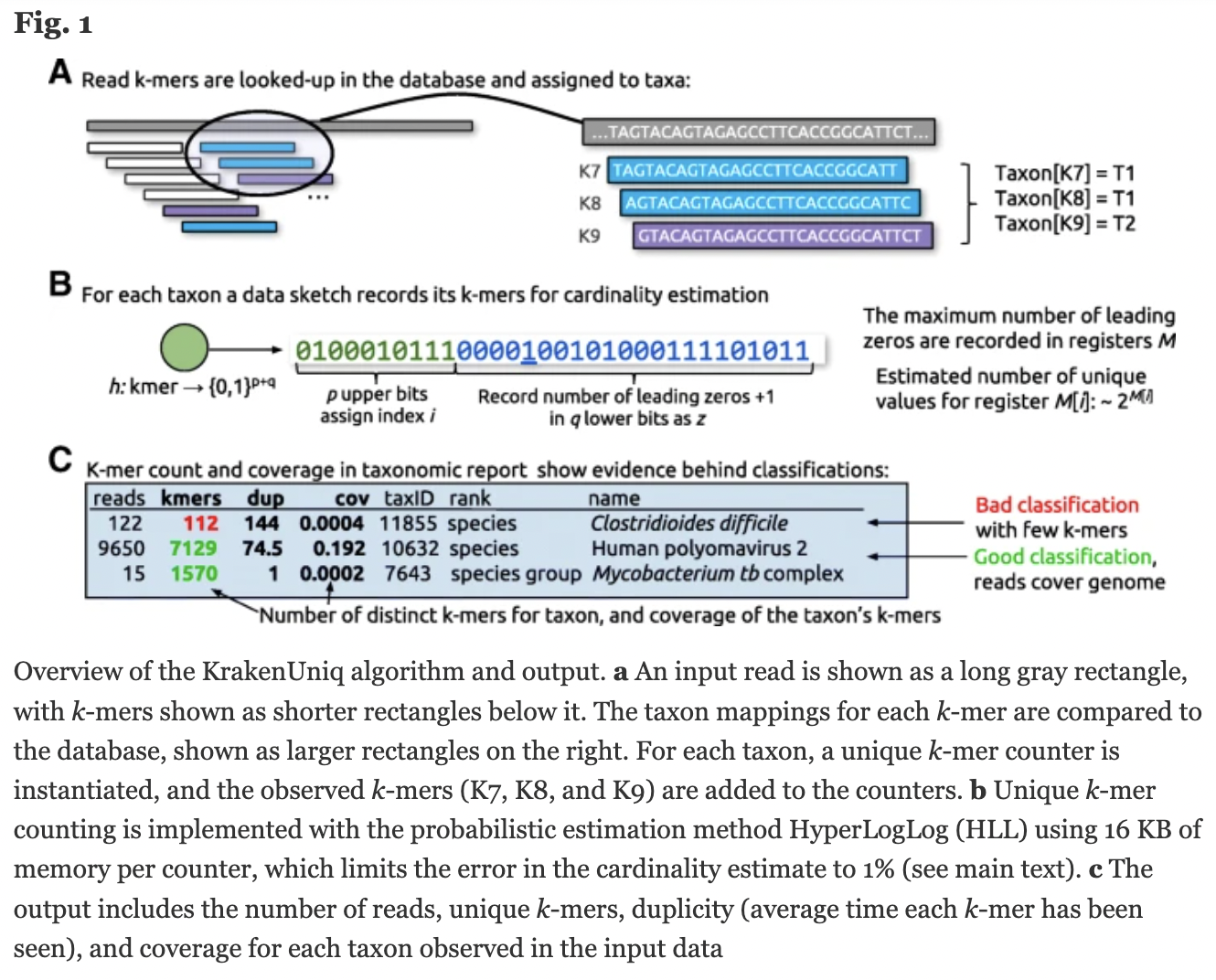
KrakenUniq gives you k-mer coverage information, reporting the number and percentage k-mers in an organism that were hit by reads. This helps to differentiate between false positives and good classifications.

https://www.biorxiv.org/content/10.1101/2022.06.01.494344v1.full.pdf
http://ccb.jhu.edu/software/choosing-a-metagenomics-classifier/#:~:text=However%2C%20while%20Kraken%20provides%20only,1%20databases%2C%20not%20Kraken%202.
https://github.com/fbreitwieser/krakenuniq/blob/master/MANUAL.md
9.2 KrakenUniq databases
KrakenUniq requires Kraken1 not Kraken2 databases
We won’t build a database since it can take several days, but let’s look at the documentation:
https://github.com/fbreitwieser/krakenuniq#database-building
You can also get prebuilt databases:
https://benlangmead.github.io/aws-indexes/k2
We will use a bacterial databases located here:
/home/data/metagenomics-2310/krakenuniq
Make a working directory and go into it.
The taxa it contains are here: /home/data/metagenomics-2310/krakenuniq/library/bacteria/library_headers.orig
Take a look at the file using head.
Click for Answer
Click for Answer
Note: this doesn’t strictly give us the number of genera as some genera are written as Frankia_* and are counted independently even though they are all part of the Frankia genus.
Click for Answer
9.4 Run KrakenUniq
Make sure you are in a screen. You can open a new screen or re-enter a screen you already have.
Activate the KrakenUniq environment.
Run Kraken on sample 3469-3.
krakenuniq \
--db /home/data/metagenomics/krakenuniq \
--threads 32 \
--report-file 3469-3.report \
--unclassified-out 3469-3.unclassified.fq \
--classified-out 3469-3.classified.fq \
~/microbe_fastq/3469-3.microbe.fq.gz \
> 3469-3.krakenuniqoutputMake sure you got the following files:
3469-3.classified.fq
3469-3.krakenuniqoutput
3469-3.report
3469-3.unclassified.fq
This command should show you all of them and give you their sizes so you can make sure none of them are empty.
Use the less command to look at each of them.
Take a closer look at the report and make sure that you understand each column (see below).
%
percent reads belonging to that taxa (including its decsendants)
reads
reads belonging to that taxa (including its descendants)
taxreads
reads assigned to this taxonomic level (and not to its descendants)
kmers
number of unique kmers
dup
average kmer count
cov
coverage of the kmers of the database clade
How many classified and unclassified reads are there?
Note: there are many ways to do this and you should get the same numbers no matter which file you look at.
What percentage of the reads are from the Frankia genus?
Click for Answer
What is the next most frequent genus? What percentage of the reads belong to this genus?
9.5 Pavian
Copy the reports to your computer.
Let’s open them in Pavian. You
If you haven’t installed Pavian, visit the installs section above.
Once you have Pavian installed, in your local Rstudio, type:
pavian::runApp(port=5000)
You can load more than one sample by shift-clicking when you choose files to load.
More information on things you can do in Pavian is in the Centrifuge chapter.
Deactivate the environment