Chapter 7 KrakenUniq
“confident and fast metagenomics classification using unique k-mer counts”
Default kmer = 31
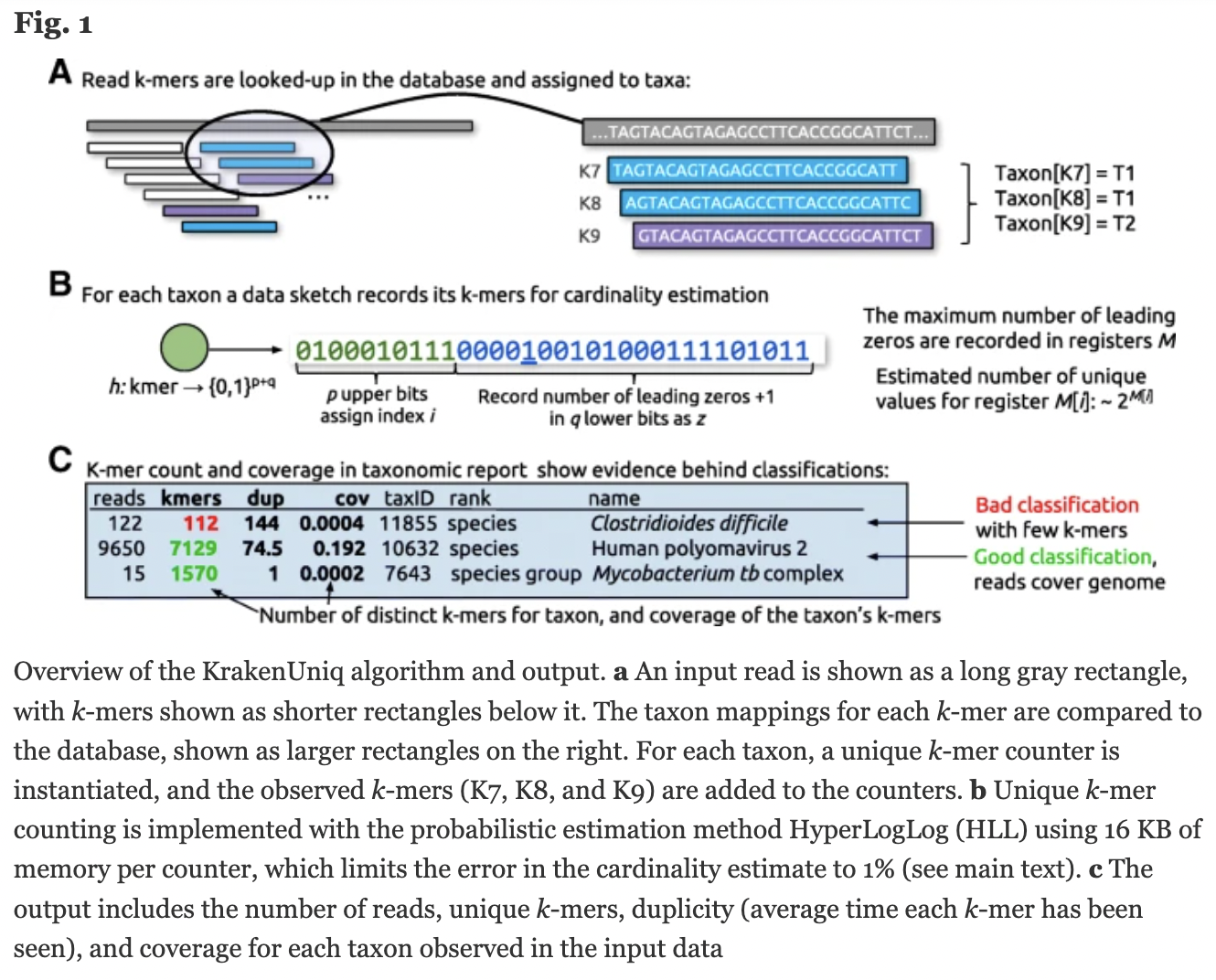
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1568-0
https://github.com/fbreitwieser/krakenuniq
https://gitlab.umiacs.umd.edu/derek/krakenuniq/-/blob/master/MANUAL.md
7.1 Kraken/Centrifuge Family
Kraken 1 is obsolete.
Centrifuge was written to improve Kraken’s memory issues. It is completely new code with a different classification index. Its databases are also much smaller. Centrifuge can assign a sequence to multiple taxa. We’ll use centrifuge later in the course.
KrakenUniq is based on Kraken 1. Adds an efficient algorithm to assess coverage of unique kmers, running at the same speed and with only slightly more memory than Kraken 1. KrakenUniq distinguishes low abundance organisms from false positives.
Kraken 2 uses “probabilistic data structures” to reduce memory and increase speed at the expense of lower accuracy that leads to false positives during the classifications (“a few dozen out of millions”).
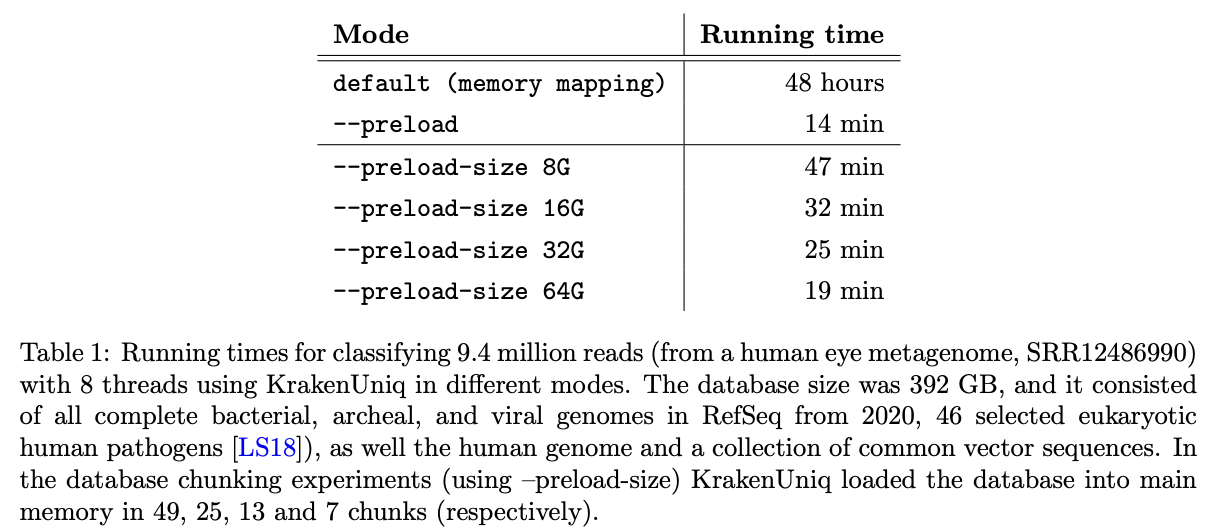
KrakenUniq was updated in May 2022 (v0.7+) that helped to deal with the large databases on computers without enough RAM to load them into memory. Databases can be read in chunks that fit in RAM.
KrakenUniq gives you k-mer coverage information, reporting the number and percentage k-mers hit by reads. This helps to differentiate between false positives and good classifications.

https://www.biorxiv.org/content/10.1101/2022.06.01.494344v1.full.pdf
http://ccb.jhu.edu/software/choosing-a-metagenomics-classifier/#:~:text=However%2C%20while%20Kraken%20provides%20only,1%20databases%2C%20not%20Kraken%202.
https://github.com/fbreitwieser/krakenuniq/blob/master/MANUAL.md
7.2 KrakenUniq databases
KrakenUniq requires Kraken1 not Kraken2 databases
We won’t build a database since it can take several days, but let’s look at the documentation:
https://github.com/fbreitwieser/krakenuniq#database-building
You can also get prebuilt databases:
https://benlangmead.github.io/aws-indexes/k2
We will use a bacterial databases located here:
/home/data/metagenomics-2310/krakenuniq
Link to it.
The taxa it contains are here: /home/data/metagenomics-2310/krakenuniq/library/bacteria/library_headers.orig
Take a look at the file. How many sequences are there?
How many genera?
awk '{print $2}' /home/data/metagenomics-2310/krakenuniq/library/bacteria/library_headers.orig | sort -u| wc -lAnd how many of each genera?
7.3 Run KrakenUniq
Make a working directory and go into it.
Activate the KrakenUniq environment.
Run Kraken on sample 3469-3.
time krakenuniq \
--db /home/data/metagenomics-2310/krakenuniq \
--threads 32 \
--report-file 3469-3.report \
--unclassified-out 3469-3.unclassified.fna \
--classified-out 3469-3.classified.fna \
~/microbe_fastq/3469-3.microbe.fq.gz \
> 3469-3.krakenuniqoutputMake sure you got the following files: 3469-3.classified.fna 3469-3.krakenuniqoutput 3469-3.report 3469-3.unclassified.fna
This command will also bring up the input file as well (3469-3.microbe.fq.gz)
Use the less command to look at each of them.
Now run it on the other samples.